Research
Overview
- In this page, I highlight my research on how machine learning and computational models enable understanding, mapping, and predicting cellular biology.
- Before 2018, as part of my AI program master thesis, I developed Deep Packet, the first neural network architecture for network traffic classification, and it has become one of the seminal papers in the field.
- Papers selected as cover:


- See a full list of papers on Google Scholar.
Generative modeling of cellular responses to perturbations
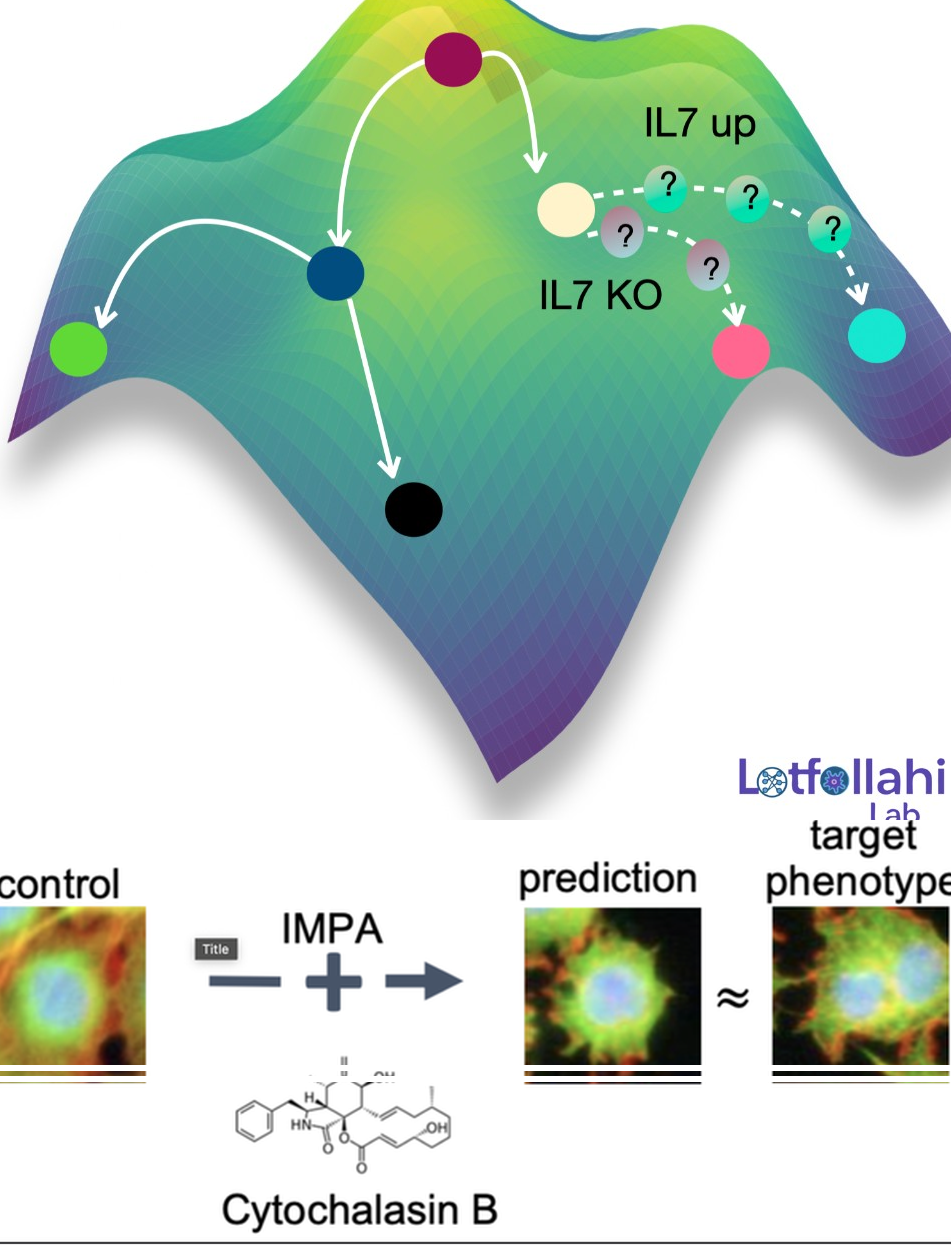
We develop generative AI models that predict how cells change in response to perturbations—such as drugs, CRISPR knockouts, disease context, or other interventions—enabling counterfactual biology: given a cell measured in one condition, what would it look like under another? This research direction has evolved into what we now call the Virtual Cell: a modeling framework aimed at learning reusable representations of cell state and using them to predict cellular responses to perturbations, including out-of-distribution generalization to unseen perturbations and combinations. The long-term goal is to turn perturbational datasets into a predictive engine for hypothesis generation, mechanism discovery, and prioritization in large-scale screens.
Across single-cell omics and high-content microscopy, we introduced a series of generative approaches that learn how perturbations transform cellular state and phenotype. Our early work scGen models perturbation effects with simple latent-space arithmetic. We then developed trVAE, which reframes perturbation prediction as distribution matching—moving cells from a control distribution to a perturbed condition. Later, during my time at Facebook AI, we introduced the Compositional Perturbation Autoencoder (CPA) to predict combinatorial perturbations such as drug combinations or double CRISPR knockouts, and extended it to handle unseen drugs (chemCPA). In parallel, we developed IMPA (Image Perturbation Autoencoder) to predict perturbation-induced morphological responses in high-content microscopy using untreated cells as input, addressing the challenge that only a small fraction of perturbations show measurable activity in experimental screens. More recently, we introduced CellDISECT, where we introduce multiple counterfactuals for disentangling covariates and predicting cellular responses.
Selected papers:
- scGen — Nature Methods (2019) — [paper], [code], [press]
- trVAE — Bioinformatics (2020) — [paper], [code], [talk]
- CPA — Molecular Systems Biology (2023) — [paper], [code], [blogpost]
- chemCPA — NeurIPS (2022) — [paper], [code]
- IMPA — Nature Communications (2025) — [paper], [code]
- CellDISECT — bioRxiv (2025) — [paper], [code]
Learning tissue architecture and reprogramming tissue ecosystems with spatial genomics and generative AI
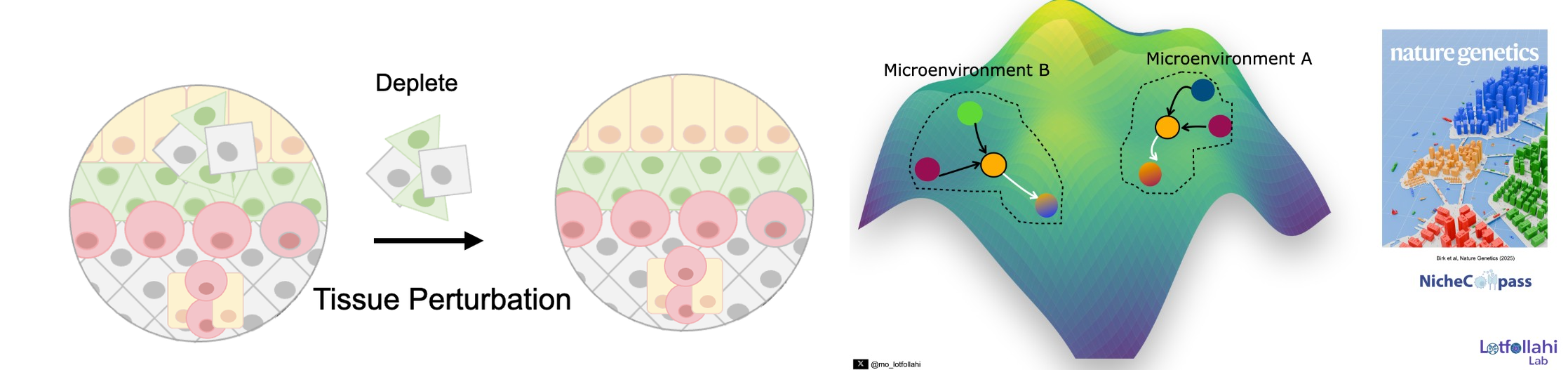
Spatial genomics makes it possible to study tissue organization at cellular resolution, but two fundamental challenges remain. First, can we identify tissue microenvironments (niches) and quantitatively define what makes them distinct—based on the cellular pathways and communication programs that structure them? Second, can we move beyond description and simulate perturbations of tissue microenvironments to predict how local ecosystems and cellular states reprogram in response to interventions?
To address the first challenge, we developed NicheCompass, a pathway-informed graph deep learning framework grounded in principles of cellular communication. NicheCompass learns interpretable representations that capture signaling and interaction programs, enabling robust niche discovery and quantitative characterization across spatial samples and disease contexts.
To address the second challenge, we developed MintFlow, a generative AI approach that learns how local tissue microenvironments imprint and reprogram cellular state, and enables in silico tissue perturbations—for example, depleting or replacing specific cell populations and predicting resulting responses at micro- and macro-scales. MintFlow shifts spatial analysis from descriptive mapping toward predictive simulation of tissue ecosystem reprogramming, supporting unbiased mechanism discovery and translational hypothesis generation across human disease settings.
Selected papers:
Machine learning to build single-cell atlases
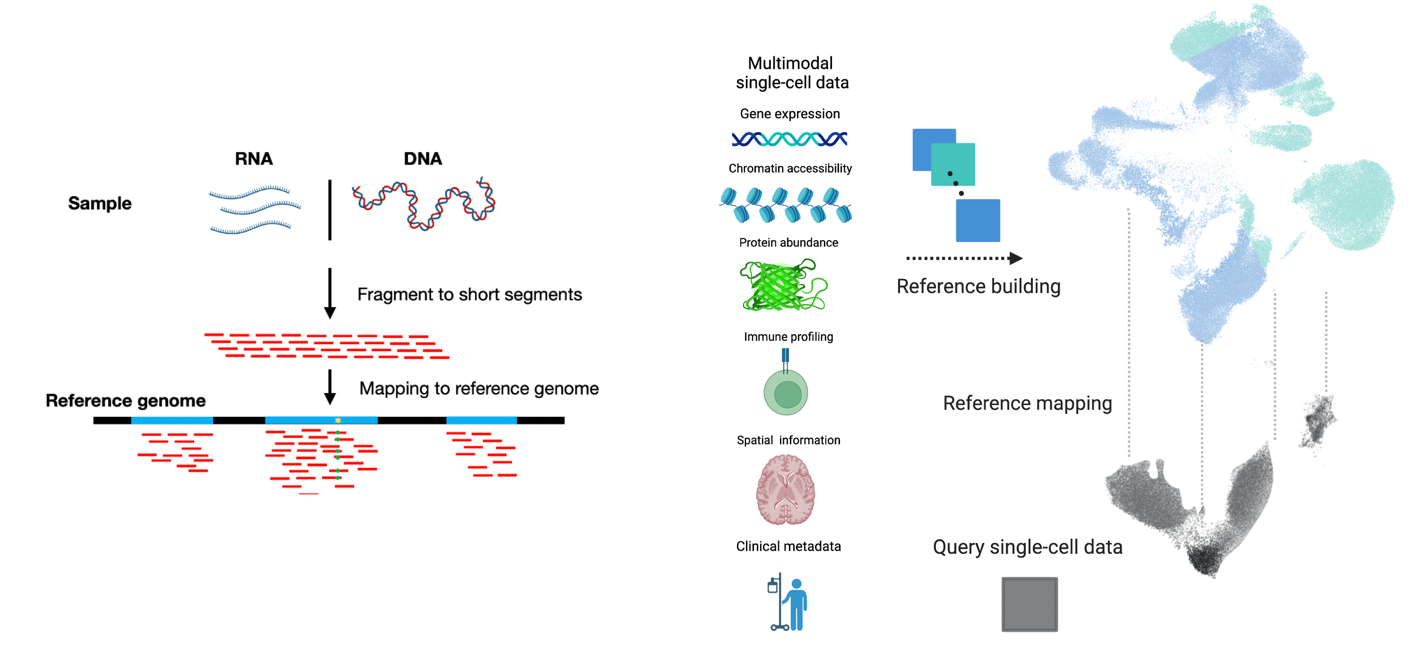
Our lab has pioneered machine learning methods that make single-cell atlases reusable, extendable references—so new datasets can be mapped, annotated, and compared in a shared coordinate system rather than re-integrated from scratch.
We introduced scArches for reference mapping by transfer learning, enabling query datasets to be mapped into a pretrained atlas without retraining the reference. We extended this to evolving cell identity structure with treeArches (updating cell-type hierarchies). We expanded atlas building beyond RNA with multimodal generative models: Multigrate integrates partially overlapping modalities (RNA/ATAC/protein) into unified multimodal references, and mvTCR integrates TCR sequences + scRNA-seq for immune atlases. To improve biological meaning and scale, we developed expiMap for interpretable gene-program embeddings and scPoli for population-level references that learn sample- and cell-level structure and provide uncertainty-aware annotation of new data.
Community impact (examples)
The community has extensively leveraged these approaches. Here are examples that our lab contributed to, showcased in building the first integrated reference atlas for lung, and more recently integrated spatial atlases for fibroblasts and skin.